Table of Contents
CEST data evaluation tool
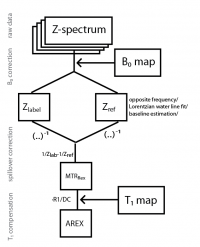 This tool allows you to evaluate CEST MRI data and create CEST images and ROI evaluations from basic contrasts as well as sophisticated multi-Lorentzian fits
This tool allows you to evaluate CEST MRI data and create CEST images and ROI evaluations from basic contrasts as well as sophisticated multi-Lorentzian fits
Dowload zipped Matlab implementations here or find the package on https://github.com/cest-sources/CEST_EVAL/
Tutorial
These routines allow you to
- import raw data
- Calculate B0 maps from Z-spectra
- B0 correct Z-spectra
- investigate CEST spectra for pixels and ROIs
- calculate contrasts (MTRasym, MTRRex, Lorentzian difference, AREX)
- fit multi-Lorentzian fits to Z-spectra
MTRRex,AREX: Zaiss et al. NMR Biomed., 27: 240–252. doi: 10.1002/nbm.3054.
Fitting: Windschuh et al. (2015), NMR Biomed., 28(5):529-37. doi: 10.1002/nbm.3283.
WASABI B1/B0 mapping and correction: Schuenke et al. (2016), MRM doi: 10.1002/mrm.26133.
1/2 Basic Z-spectra evaluation
2/2 Lorentzian fitting for isolated CEST evaluation
There are 4 important inputs:
- M0_stack(x,y,z) the thermal M0 magnetization, or the normalization image w/o saturation
- Mz_stack(x,y,z,w) the stack of saturated images at frequency w
- dB0_stack_int(x,y,z) the internal dB0 map in ppm calculated by the minimum of Mz_stack
- dB0_stack_ext(x,y,z) an external dB0 map in ppm
further we need the frequency offset axis, or the omega-axis (w-axis) of the Z-spectrum. It is saved in the parameter struct P under the sequence parameters P.SEQ
- P.SEQ.w
To calculate the internal B0 map the function MINFIND_SPLINE_3D is called after that the B0-correction and normalization can be performed and the final stack is
- Z_corrExt - the B0-corrected and normalized stack of Z-spectra
Alternatives
MITK
The Medical Imaging Interaction Toolkit (MITK) is a free open-source software system for development of interactive medical image processing software. MITK combines the Insight Toolkit (ITK) and the Visualization Toolkit (VTK) with an application framework. As a toolkit, MITK offers those features that are relevant for the development of interactive medical imaging software covered neither by ITK nor VTK, see the Toolkit Features for details. It recently got an extension for CEST analysis MITK CEST:
JHU
find the CEST data processing tool of the John Hopkins group here
